New VAERS PRR Interactive Dashboard Introduction
The "How To" guide...
https://public.tableau.com/app/profile/alberto.benavidez/viz/VAERSC19vsAllElse20250915/Home
Most people do not realize the “symptoms” that every pharmacovigilance system in the world uses are MedDRA codes. Standardized terms and identifiers within the Medical Dictionary for Regulatory Activities (MedDRA), is a clinically validated international medical terminology dictionary-thesaurus developed under the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH). It is primarily used by regulatory authorities, pharmaceutical companies, and healthcare professionals to classify and report adverse events, medical conditions, medicines, and medical devices in a consistent, harmonized manner across global pharmacovigilance and clinical development activities. Key Features and Structure MedDRA is organized hierarchically to enable precise data entry, retrieval, and analysis:
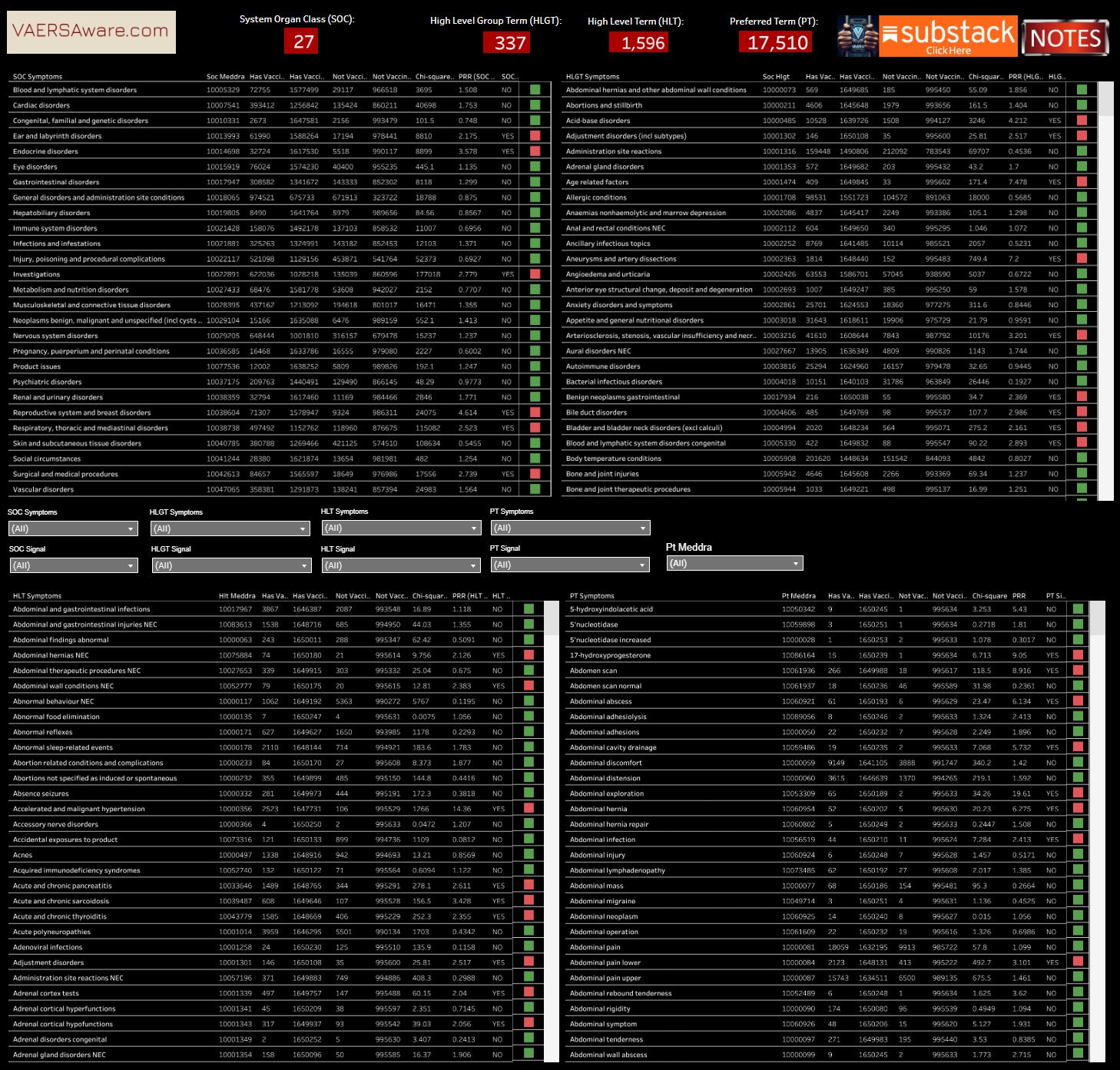
System Organ Class (SOC): The highest level, representing broad categories like anatomical or physiological systems (e.g., "Gastrointestinal disorders" or "Infections and infestations"). There are 27 SOCs.
High-Level Group Term (HLGT): Subdivisions within SOCs for more specific groupings.
High-Level Term (HLT): Further refinements of HLGTs.
Preferred Term (PT): A specific, non-classifiable descriptor for a medical condition or event (e.g., "Nausea" as a PT).
Lowest Level Term (LLT): The most granular level, synonymous with or more detailed than PTs, used for data entry.
Additionally, MedDRA includes Standardized MedDRA Queries (SMQs), which are pre-defined groupings of terms to aid in detecting potential safety signals, such as "Myocardial ischaemia" or "Anaphylactic reaction."
Purpose and Usage:
MedDRA facilitates the sharing of safety data internationally, supporting regulatory submissions, post-marketing surveillance, and clinical trials.
It is subscription-based and updated biannually (e.g., versions like 21.0 or later) to reflect evolving medical knowledge.
In many countries, its use is mandated for electronic reporting of adverse events to agencies like the FDA or EMA.
Beyond pharmaceuticals, it's increasingly applied in other sectors like medical devices, vaccines, and even non-drug industries (e.g., cosmetics or tobacco) for capturing health events.
MedDRA emphasizes multi-axial concept representation, meaning terms can link across multiple SOCs for comprehensive analysis without losing data integrity. For official access or training, refer to the MedDRA website (meddra.org). These are not ICD-10 codes.
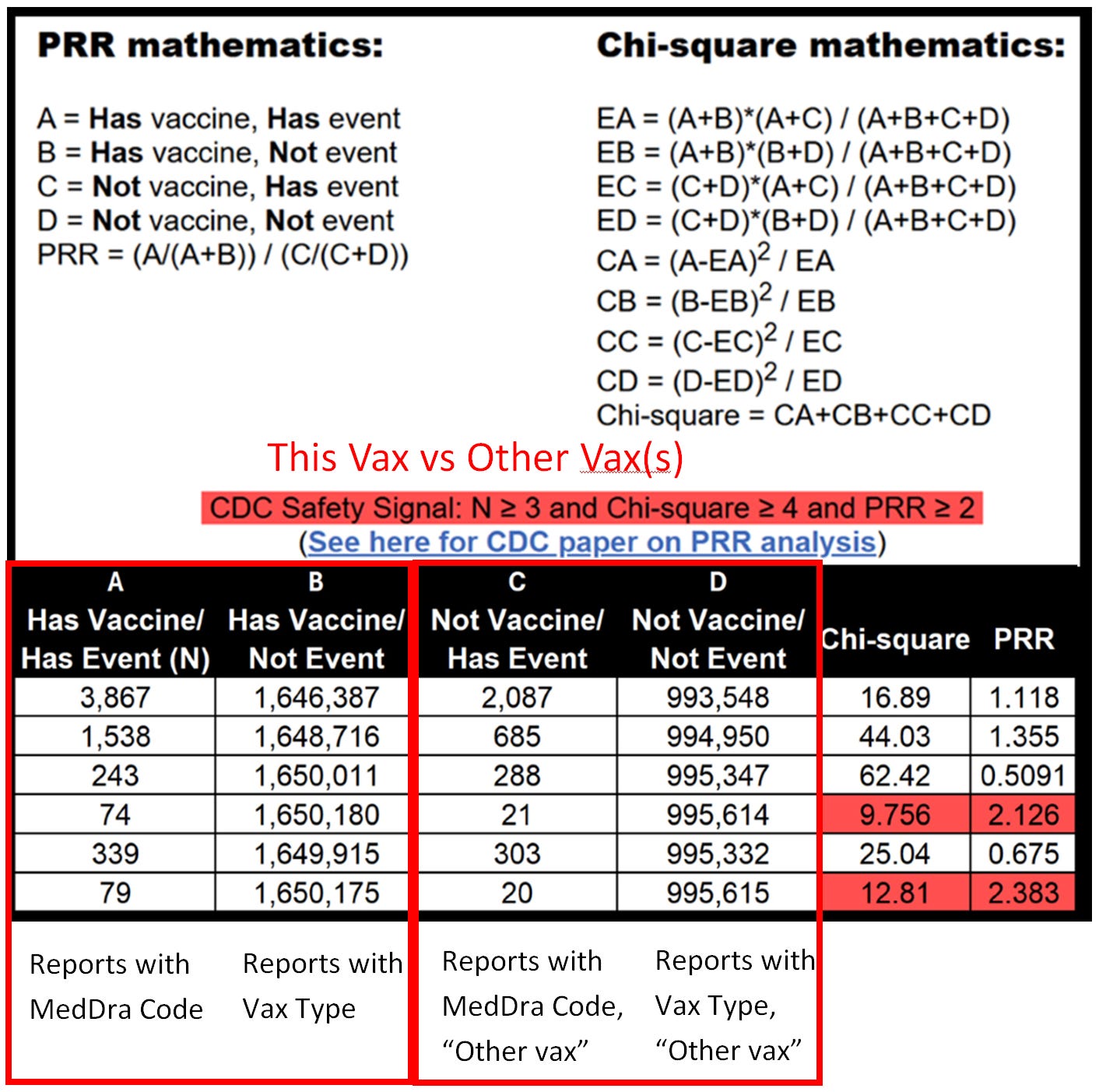
My last article explains the math for the proportional reporting ratio (PRR).
This Dashboard maps the PRR through each MedDRA level from Preferred Term (PT) to System Organ Class (SOC). The image above is the medalerts.org example of how to interpret the structured data within medalerts.org native environment and this dashboard.
Disclaimer: Medalerts.org PRR tool is very powerful and can allow for an infinite amount of queries and combinations of analysis. Our interactive is not currently as dynamic in query options. This dashboard is simply visualizing the highest level calculations on Covid-19 jabs versus on all other vax types, minus UNKNOWN vax type. It must be recognized that any unknown vax type “co-mingled” into any unique report/case then gets included (unfortunately).
Here is an example of PRR at the SOC level:
Do you see what the #1 SOC is? This is what I was screaming about long before anyone else when I noticed heavy menstruation, miscarriages and fetal demies was being under represented looking at PRR for just “severe event” levels. The simple reason is most miscarriages and stillbirth for that matter do not rise to the level of in-patient hospitalization or even a Life Threatening event. Here’s an old meme I created in 2021:
Here is why they can not see a death signal!
No wonder why CDC/FDA/VAERS does not put an applicable “death” MedDRA code on every valid death report, because only around ~half the legitimate covid jab deaths actually have a legitimate “death” medDRA code!
Let’s suppose we could make an honest PRR calculation on death, it would look more like this:
Grok is pretty cool:
To compute the Proportional Reporting Ratio (PRR) and Chi-square statistic for the given data, we use the standard pharmacovigilance formulas for signal detection. The data provided is:
COVID-19 vaccine:
Death reports: 38,773
Total adverse event reports: 1,656,122
Non-COVID-19 vaccine:
Death reports: 11,030
Total adverse event reports: 1,012,027
Step 1: Construct the 2x2 Contingency Table
Non-death for COVID-19 = 1,656,122 - 38,773 = 1,617,349
Non-death for non-COVID-19 = 1,012,027 - 11,030 = 1,000,997
Grand total = 1,656,122 + 1,012,027 = 2,668,149
Step 2: Calculate PRR
PRR = [ (Death reports for COVID-19) / (Total reports for COVID-19) ] / [ (Death reports for non-COVID-19) / (Total reports for non-COVID-19) ]
= (38,773 / 1,656,122) / (11,030 / 1,012,027)
≈ 0.02341 / 0.01090
≈ 2.147
Step 3: Calculate Chi-square Statistic
The Chi-square test assesses whether the proportions of death reports differ significantly between COVID-19 and non-COVID-19 vaccines.
Expected values for each cell:
Expected deaths for COVID-19 = (1,656,122 × 49,803) / 2,668,149 ≈ 30,919.74
Expected non-deaths for COVID-19 = (1,656,122 × 2,618,346) / 2,668,149 ≈ 1,625,202.26
Expected deaths for non-COVID-19 = (1,012,027 × 49,803) / 2,668,149 ≈ 18,883.26
Expected non-deaths for non-COVID-19 = (1,012,027 × 2,618,346) / 2,668,149 ≈ 993,143.74
Chi-square = Σ [(Observed - Expected)² / Expected] for all cells:
= [(38,773 - 30,919.74)² / 30,919.74] + [(1,617,349 - 1,625,202.26)² / 1,625,202.26] + [(11,030 - 18,883.26)² / 18,883.26] + [(1,000,997 - 993,143.74)² / 993,143.74]
≈ 2,011.74 + 3.80 + 3,287.84 + 6.22
≈ 5,309.60
Interpretation
PRR = 2.147: A PRR > 2 suggests a higher proportion of death reports for COVID-19 vaccines compared to non-COVID-19 vaccines, potentially indicating a signal in pharmacovigilance (though PRR alone doesn't confirm causation).
Chi-square = 5,309.60: With 1 degree of freedom, this is highly significant (p-value ≈ 0), indicating a statistically significant difference in death reporting proportions between the two groups.
Note: VAERS data reflects unverified reports and is subject to biases (e.g., reporting rates, vaccination volume). A PRR > 2 and significant Chi-square suggest a disproportionate reporting signal, but further investigation is needed to assess causality or clinical significance.
The Eagle’s note of the note, the only biases we are being subjected to here is nefarious intentions of VAERS Administrators. Not to mention the additional ~800 extra covid jab deaths in the follow-up reports that are not even being considered in these calculations. God Bless
A very encouraging message from my friend Dr. David Wiseman:
This is a very tricky subject, even before we get to the VAERS shenanigans you have been documenting.
I have a discussion in my paper about why the PRR threshold is 2, and why the chi square magic number is 4. Because of various statistical quirks, chi squared is not a good idea, and the "standard" needs to use a confidence interval approach. Evans, in his original paper was not rigid in using chi-squared, and suggested confidence intervals could be used.
This is all in addition to setting the threshold properly and adjusting for masking.
As for multiple vax dosing, (and assuming it is all reported properly?!) this is certainly something for a nice feature of VAERS wonder to be able to explore.
Thanks again
best
DW










This is great stuff because you've been the only one telling us for years that, in reality,
"The “Symptoms” That EVERY Pharmacovigilance System In The World Uses Are MedDRA Codes."
That's the best, me, a lay person, can decipher. And you're trying to translate to us, how that works for everything in the medical field, and particularly how it "works" for VAERS. Right?
I feel like I'm trying to read Chinese.
Thank you for giving us the MedDRA alphabet.
(Still going to email and verify your mailing, just falling behind this month).
Proof of the old saying that figures don't lie but liars figure.. bless you Albert!! <3